
Images and movies (unless otherwise noted) are free for educational use under a
Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License.
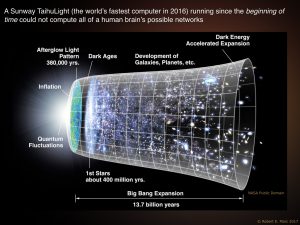 You cannot compute a brain. You cannot compute a brain.
|
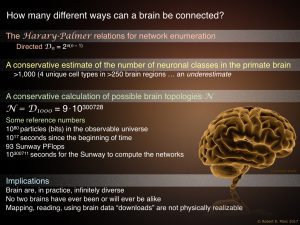 How complex are brains? Graph Theory. How complex are brains? Graph Theory.
|
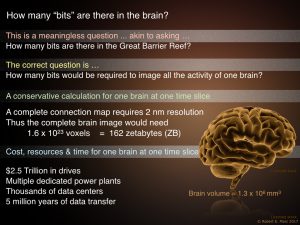 How many bits are there in a brain? How many bits are there in a brain?
|
 Why can’t we download our brains? Why can’t we download our brains?
|
 Why should we invest in brain research? Why should we invest in brain research?
|