| Micromolecules: definitions & links | Cellular domains: Genome, preoteome & metabolome |
| Metabolic diversity: Scale, dynamics, & phyletics | Phenotyping strategies: proteomics vs metabolomics |
| CMP Platforms: Platforms and workflow overview | CMP Probes: The probe library |
| CMP Substrates: Molecular trapping & detection | CMP Datasets: Data arrays for multichannel imaging |
| CMP Analysis: pattern recognition theory and tools | CMP Exploration: N-space visualization tools |
| CMP Annotation: browsing & annotating data |
Micromolecular Diversity
The Scale of Metabolic Diversity:
Most of us learned intermediary metabolism via the general model of bacterial or perhaps hepatocyte biolochemistry. Two key features were missing: real metabolite concentrations and assessments of metabolic diversity across cell types. The analysis of physiological interactions such as nutrition, signaling, growth regulation, and defense mechanisms in complex heterocellular tissues requires a clear understanding of the metabolomes of each system compartment. Metabolic modular composition of tissues is visualized by CMP, even when gene or protein expression patterns cannot be invoked to detect them. CMP shows that metabolic variation across cell types is extreme and multidimensional. The scale of diversity ranges from fine and discrete to broad and continuous. This is is reflected across in the modular organization ranges across:
- complex gridded cell mixtures that form 2D (Fig 1, retina) or 3D (cortical) modules
- large-scale functional modules where each module is composed of many cells (Fig 2, renal cortex)
- complex non-gridded cell mixtures such as pancreatic islets
- functional gradient systems of coupled cells (Fig 3, liver modules).
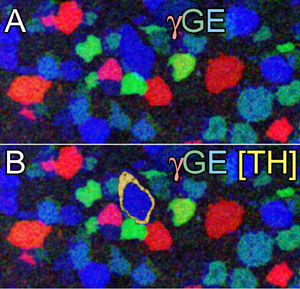
Fig 1 Three-channel mapping of (A) GABA (gamma = red), glycine (g = green), and glutamate (E = blue) signals in registered ultrathin sections of rabbit retinal amacrine cells. (B Four-channel mapping by addition of tyrosine hydroxylase (TH) signals as a yellow alpha channel. Image width = 120 µm.

Fig 2 CMP imaging of free amines in mouse renal cortex.tt.J.E = r.g.b mapping (tt taurine, J glutathione, E glutamate. D distal convoluted tubules, G glomerulus, GTH glutathione-rich epithelia, K thick Henle’s tubules, N thin Henle’s tubules, P proximal convoluted tubules, VP vascular pole, VR vasa recta, UP urinary pole, US urinary space. Image = 256 × 360 µm.

Fig 3 CMP visualization of metabolic gradients in hepatic modules: Q.E.J = r.g.b mapping (Q glutamine, E glutamate, J glutathione. C central vein, P portal vein. Image size 346 × 253 µm. Inset is the glutamine signal around the module central vein; inset size 35 × 33 µm.Glutamine levels drop 2.5× and glutamate levels rise 10× over the path defined by the arrow.