| Micromolecules: definitions & links | Cellular domains: Genome, preoteome & metabolome |
| Metabolic diversity: Scale, dynamics, & phyletics | Phenotyping strategies: proteomics vs metabolomics |
| CMP Platforms: Platforms and workflow overview | CMP Probes: The probe library |
| CMP Substrates: Molecular trapping & detection | CMP Datasets: Data arrays for multichannel imaging |
| CMP Analysis: pattern recognition theory and tools | CMP Exploration: N-space visualization tools |
| CMP Annotation: browsing & annotating data |
Cytosomic Domains
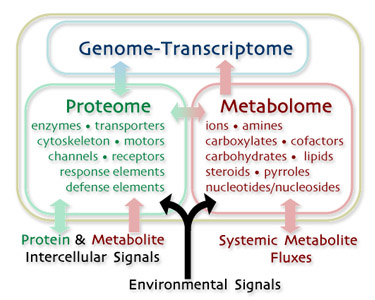
Phenotyping cells requires tools to probe the separate but interacting domains of cellular identity.
The Genome/Transcriptome is the set of realized cellular instructions (nucleotide strings): the operating system.
The Proteome is the set of realized cellular devices (folded amino acid strings): the application code.
The Metabolome is the set of realized cellular diffusion-reaction chemistries: real-world data processing events.
The Cytosome is the union of these sets.
Cytosomics is a superplatform of technologies designed to harmonize multichannel datasets of gene, protein and metabolite expression profiles with single-cell resolution and comprehensive coverage. In practice, a functional genomic signature is realized by tracking mRNA signals in known cell types. Gene function is amplified 2-5 decades as protein arrays and a proteomic signature is a snapshot of protein macromolecular signals. Protein functions are amplified 3-6 decades as molecular fluxes. This dynamic metabolomic signature is a mix of micromolecular signals as diverse as the protein device array that shapes it.
These stages of amplification are reflected in the visualization strategies required to detect genomic, proteomic and metabolomic targets. Many micromolecules are so abundant (0.1-10 mM) that they can be probed in pure surface assays. Most cytoplasmic proteins are expressed at lower levels (10-1000 nM), requiring optical superposition to achieve reliable detection. Finally, mRNA signal detection requires long integration times. A fusion of detection platforms is required to acquire comprehensive cytosomic signatures.