METHODS: Plastic embedding
Plastics
Epoxy resins are optimal matrices for HPI tm: High Performance Immunocytochemistry. The physical attributes of modern “plastic” support matrices emerged from the needs of electron microscopy – thermal stability, resistance to deformation, low mass thickness, low viscosity, etc. The epoxy resins, whose properties are influenced by the ratio of cross-links to end-links, have dominated the electron microscopic field. The properties that render them physically “tunable” for electron microscopy also make it possible to etch the resin away, exposing hapten sites for attack by IgGs. Weak etching is employed in electron microscopic immunocytochemistry to allow partial antigen exposure while retaining sufficient section rigidity in the electron beam. For HPI tm, epoxy resins allow cutting of extremely thin sections for serial superposition and can be completely etched (deplasticized) to provide optimum antigen exposure and IgG attack. This regime of accessible antigen is generally less than ten nanometers in depth, as large IgGs cannot penetrate the mesh of cross-linked tissue effectively. Thus signal strength is largely independent of section thickness. Alternatives such as Araldite resins or epoxy/Araldite mixtures designed to improve section rigidity for electron microscopy do not etch as completely as pure epoxy resins and, while usable, provide less antigen mobility. The more hydrophilic methacrylates have gained popularity in post-embedding macromolecule immunocytochemistry and provide some hapten access. As they must be used without etching, however, they yield substantially weaker hapten signals. Conventional paraffin and some newer hard paraffins permit reasonably thin sections for immunocytochemistry and, after clearing, yield quite good signals. The key limitations with such matrices are their excessive thicknesses and incompatibility with electron microscopy.
Stacks and Mosaics
A laminar assembly of several samples into one bloc is a stack. A single section from a stack, however thin it may be, contains representatives of every sample which can then be probed and analyzed under identical conditions. Stacks are simple to construct and the ability to section 12 or more specimens simultaneously more than compensates for the day or two spent constructing the stacks. A mosaic is an arrangement of several pieces of tissue side-by-side on a pre-existing bloc face, permitting simultaneous sections of material not necessarily compatible with a stack assembly. Mosaics require substantially more care in preparation.
There are many possible methods for constructing stacks and three fundamental designs are considered here. Samples which naturally form nearly flat sheets (retina; slices of brain, liver, kidney, etc.; thin plant material such as leaves; sheets of cultured cells or cells suspended in thin sheets of agarose, and so on) may be processed for embedding in an epoxy resin and, at the final polymerization, can be flat cured under a weight:
-
-
- STACK METHOD 1
-
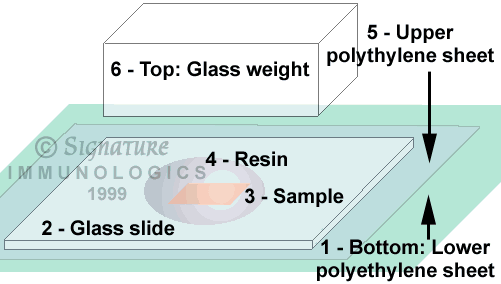
- Place the sample in 100% resin on a glass slide and cover with a sheet of 2 mil or thicker polyethylene larger than the slide. Place on a second sheet of polyethylene on a flat surface and cure under a 25 gram weight (aluminum or glass blocks).
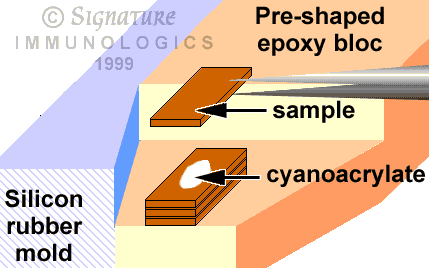
- Gently scribe 3 borders around the desired sample with a #11 scalpel blade. The very tip of the blade will break off; be sure you do not re-embed it in the stack. Scribe all the way to the glass. Cover the sample with transparent tape and scribe the 4th border. Slip the tip of the scalpel under an edge and gently pry up the isolated chip.
- Place a dot of cyanoacrylate glue about 1/3 the width of the chip on the prepared surface of a notch in the tip of a flat trapezoidal cured bloc held in its blue silicon rubber mold (source: Ted Pella) and cover with a sample. Press gently for 30 sec until the sample is fixed in place. Repeat using the first sample as the base of the 2d, and so on. Finish with a unique sample to provide orientation. Fill the notch slowly with warm resin and cure.
-
-
- STACK METHOD 2
-
- Embed tissue in epoxy resin and cure.
- Section the tissue at 30-120 um on a sliding microtome using the method of West (West R 1972 Superficial warming of epoxy blocks for cutting of 25-150 micron sections to be resectioned in the 40-90 nm range. Stain Technol 47: 201-204).
- Collect sections from different samples, trim to a common shape and embed as described in Stack Method 1.
-
-
- STACK METHOD 3
-
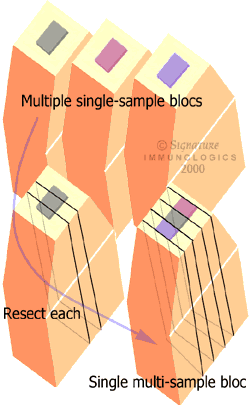
- Carefully resect slices from each source bloc with a 3X000 jeweler’s blade.
- Assemble slices into a single stack and position with cyanoacrylate.
- Coat in epoxy resin and recure.
-
-
- Mosaic Method
-
- Obtain samples of constant thickness, preferably by thick sectioning.
- Trim the sample to the smallest manageable dimensions.
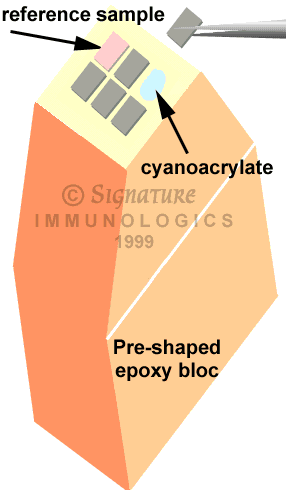
- Place a small dot of cyanoacrylate on the face of a clean bloc and gently glue the sample in place. Use the flat handle side of forceps to apply pressure if needed.
- Continue to mosaic the samples together.
- Apply a small amount of epoxy resin to the face, place in a fitted mold and recure.
Section Thickness
HPI obtains part of its precision by being insensitive to many sources of variability, such as variations in section thickness. Control of section thickness is important in obtaining precise registrations, but is not required for a good HPI signal. Post embedding labeling methods are two-phase mechanisms: IgGs partition from the fluid phase onto an essentially impenetrable surface. Even when samples are fully deplasticized, the density of cross linking in glutaraldehyde fixed material is so high that large radius of gyration molecules such as IgGs cannot penetrate. IgG binding and detection by HPI tm is thus independent of section thickness over a large range (certainly over 40-1000 nm). The practical import of this is not that one may relax the need for precision, but rather that very thin sections can be used without loss of signal strength, allowing probing of many signals in a single set of cells. Sections as thin as 40 nm have been used in HPI tm for optical microscopy (Marc, R.E., R. Murry and S.F. Basinger 1995 Pattern recognition of amino acid signatures in retinal neurons J. Neurosci. 15: 5106-5129.
Sectioning tips
As noted above, section thickness is has no little meaningful impact on signal strength in HPI tm. In practice, 200-250 nm sections are as easy to handle as routine sections >500 nm and yield substantial improvements in spatial resolution and quality of image registration. Serial sections should be cut but precision trimming to produce a ribbon is actually somewhat detrimental as HPI tmrequires the sections to be separated. Thus is it quite satisfactory to face blocs by hand rather than by glass knives. Each section should be placed on a droplet of filtered deionized water on a well of a multiwell glass slide, such as a 12 well Teflon® coated slide. One useful technique places the cells in a reverse N pattern, i.e. number the spots across the top as 1-3-5-7-9-11 and the bottom as 2-4-6-8-10-12 and place the sections down in sequence. This facilitates the placing of IgG droplets in various patterns. If you wish to probe every other section with the same IgG, then simply placing the IgG on every well of the upper or lower row will suffice. If you wish to use electron microscopic thinness sections such as silver or gold or even superthin sections, it is easiest to remove each section from the boat by floating it in a droplet within a transfer loop. Transferring the section to the slide requires care and a stereomicroscope or magnifying glasses. Gently touch the leading edge of the loop to the slide at about 30 degrees from horizontal and flex the loop with a little downward pressure to force the droplet to touch the glass. Wick the water away slowly with a small paper point (available from Ted Pella, Inc.) placed just outside the front edge of the loop and pushed into the crease between loop and glass. The section should slowly descend to the glass surface and, with luck, should remain free of folds. While tedious, use of 60 nm sections allows sampling 12 serial haptens in less than 3/4 um.
Deplasticizing
Deplasticizing epoxy resin sections involves basic attack on the end-linked epoxide rings. In electron microscopic post-embedding immunocytochemistry, strong aqueous hydrogen peroxide is used to disrupt the links but attacks proteins as well. The etching is quite limited however. For light microscopic purposes, the entire epoxy resin matrix may be removed by using anhydrous bases such as sodium ethoxide or methoxide These agents will disrupt the end-links and rapidly dissolve the polymer but, in the absence of water, will not attack most hydrophilic biological molecules. Indeed, the addition of even trace amounts of water to the solution will produce a stunning slide-cleaning solution that will remove all traces of tissue. On the whole, some proteins, peptides and many amino acids have been successfully deplasticized with these methods, restoring strong immunoreactivity.
Deosmication
Electron microscopic immunocytochemistry requires deosmication for the restoration of some immunoreactivity, the suppression of spurious gold deposition at osmium rich sites, and for light microscopic immunocytochemistry, the removal of osmium itself as it will spontaneously catalyze the silver intensification reaction and quench the gold-mediated reaction. It is not clear that osmium itself seriously impedes hapten immunoreactivity as reasonable immunogold GABA signals can be had in fully osmicated electron microscopic samples. However, osmium does seem to restrict hapten mobility which may attenuate signal strength significantly for some residues. Deosmication involves the reversal of the multiple coordination bonds formed by reoxidizing osmium with very reactive species such as periodates. Typically sodium metaperiodate or periodate are used after deplasticizing to reverse the osmication. For most small haptens, this seems fairly innocuous and full immunoreactivity of over 20 small haptens (mostly amino acids) can be restored. However, some molecules such as glutathione are strongly modified by osmication and recovery of immunoreactivity may require extensive periodate bleaching. Thus more complex species, and especially those containing thiols, are likely not always compatible with electron microscopic analysis if osmication is desired.
Disclaimer: As conditions of use are outside our control, we make no warranties, express or implied, and assume no liability in connection with use of this protocol.
© 2000 Signature Immunologics Inc. [reproduced by permission]
This protocol may be copied and distributed for educational and non-profit
purposes as long as acknowledgement of our copyright is included
in every instance of use. It may not be sold or distributed for commercial gain.