| CMP ANALYSIS |
| 1. A Clustering Tutorial for Anatomists
Introduction: Traditional statistical anatomy tests differences in metrics across cell classes. But these classes are prefigured: either they came from distinct specimens or visual classifications. As analyses of cellular populations, states, life cycles, transformations, etc. become more complex, we are faced with a difficult question: How do we classify cells? Here’s an example of the problem . A former student of mine did a wonderful Master’s thesis classifying sensory cells and found 4-6 anatomical classes in our model system. But these cells proliferate from neurogenitors, function and die. So how did we know that some of these classes weren’t life-cycle variants? We didn’t and still don’t. We had no tools for classification other than shape. But if we had other tools, how might we use them? We’d likely start with cluster analysis. |
Cluster analysis is a small branch of a larger statistical field known as pattern recognition which seeks structure or “patterns” in data metrics. Cluster analysis focuses on finding concentrations of data values. A key part of cluster analysis is finding methods that are semi-automatic. We turn them loose and they find data features. Of course it isn’t that simple, but a number of methods have been developed. One of these is the migrating or K-means method (KMM). The KMM explores a data set by:
- Selecting K initial sets of data means or centers: P(0),Q(0),R(0)….
- Assigning all data closest a center as members of that class: (class P, etc.)
- Calculating a new center value from all the data in the class, yielding P(1), Q(1), R(1)….
- Repeat 2, then 3 until the centers don’t change anymore
The trajectory of P(0) → P(1) … → P(n) is the migration of the mean. If the data are cooperative, you don’t even have to pick reasonable initial values. Cool. |
| 1D sample dataset . Here is a sample problem and KMM solution to introduce the idea more concretely.
A fluorescence dataset reports the expression of a critical molecule A in a collection of cells. It is believed that three different cell classes are present in the sample. Does the fluorescence image of A visually report that segmentation? How many cell classes does it look like to you?

The simplest strategy is to form a histogram of A values, and three overlapping modes of A thus become obvious. While cluster analysis is basically unnecessary for 1D data, it provides a good test case for the KMM.
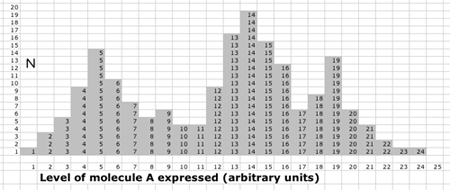
|
| 1D KMM. |
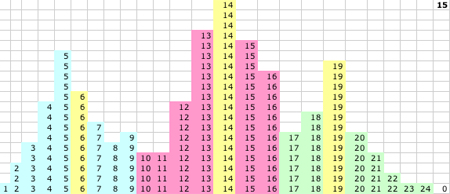
| 1D KMM Step 0. The KMM requires a single user input to begin: the number of expected classes K. The simplest implementation then picks K initial centers. Here we choose P(0) = 3, Q(0) = 5, and R(0) = 8 (yellow), but many implementations use automatic initiation. The initial task of the KMM is to assign the three classes based on Euclidian distance. Class P captures values ≤ 3 (blue) and shares half of the values between 3 and 5 (purple) with class Q. Class Q captures all values closer to 5 than 8 (red) and class R captures everything else (green).
|
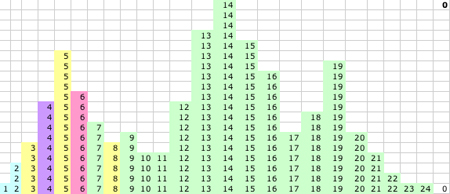
1D KMM Step 1. The KMM recalculates the K centers based on the new memberships in the classes and P(1)=3, Q(1)=5 and R(1)=15. R(1) is the only major revision at this step, but this radically changes the class memberships between R and Q.
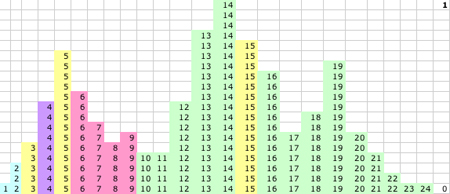 |
1D KMM Step 3. After a couple of cycles center Q starts to move.
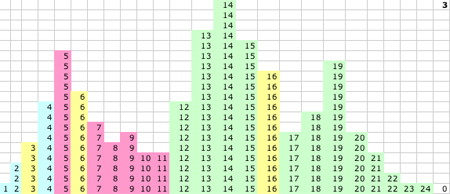 |
1D KMM Steps 0-15. The migrating means
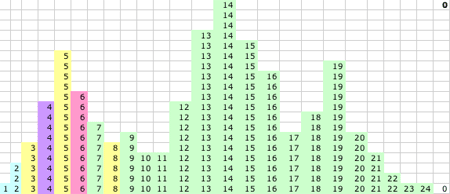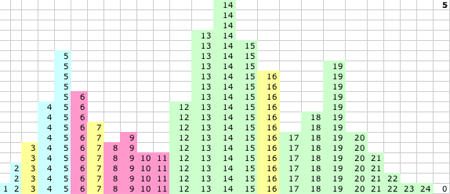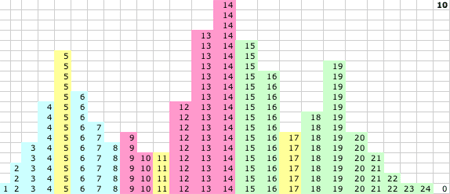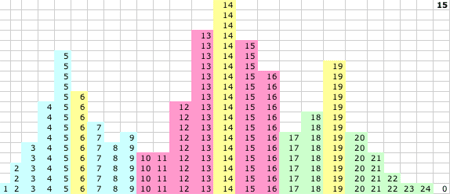 |
1D KMM Step 15. The KMM stops or converges on this solution:
- P(15) = 5.52 and P captures values 1-8
- Q(15) = 13.7 and Q captures values 10-16
- R(15) = 19.2 and R captures values 17-24
|
| 1D KMM Theme Map. Using these values, we recode the original greyscale image according to class memberships P, Q, and R.
|
1D KMM & the Normal Distribution.
- Are these memberships correct? Presume that each mode is an independent normally distributed population. The essential outcome of any scheme to classify data is a set of decision boundaries and if the correct boundaries are defined as the crossing points of the underlying distributions, the KMM is as close as one could expect from such a small sample with discrete values. But there are errors in this result, regardless of whether we use distributions or KMM to make the decision boundaries
- Where are the errors? Two problem spots are evident near the crossovers. Between classes Q and R, the overlap of the distributions suggests that any classification in this area is subject to error. In this case it is pretty large. Defining the rate of misclassifications as the ratio of half of the overlapping instances to the class size, a cell will be incorrectly grouped about 11% of the time for class Q and 19% of the time for R. Between classes P and Q, there are more data instances than predicted by the distributions. How do these cases get handled? In the 1D case, with no independent test other than the strength of A signals, there is no way out except to presume that the error rate here is similar to a real overlap rather than a fourth group of cells, with an error of about 7%.
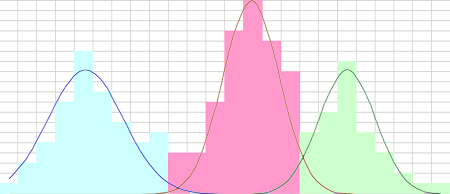
Note that the three classes are clearly drawn from different distributions and a Student’s t-test on any pair will lead to extremely small p values. There is no question of the significance of the classes, but there is a clear question of their separabilities.
- Significance: the probability that the class distributions arise from the same parent distribution.
- Separability: the probability of erroneous classification
Separability is a more stringent measure than significance. If classes are truly separable with a small degree of error, they are explicitly significant. |








