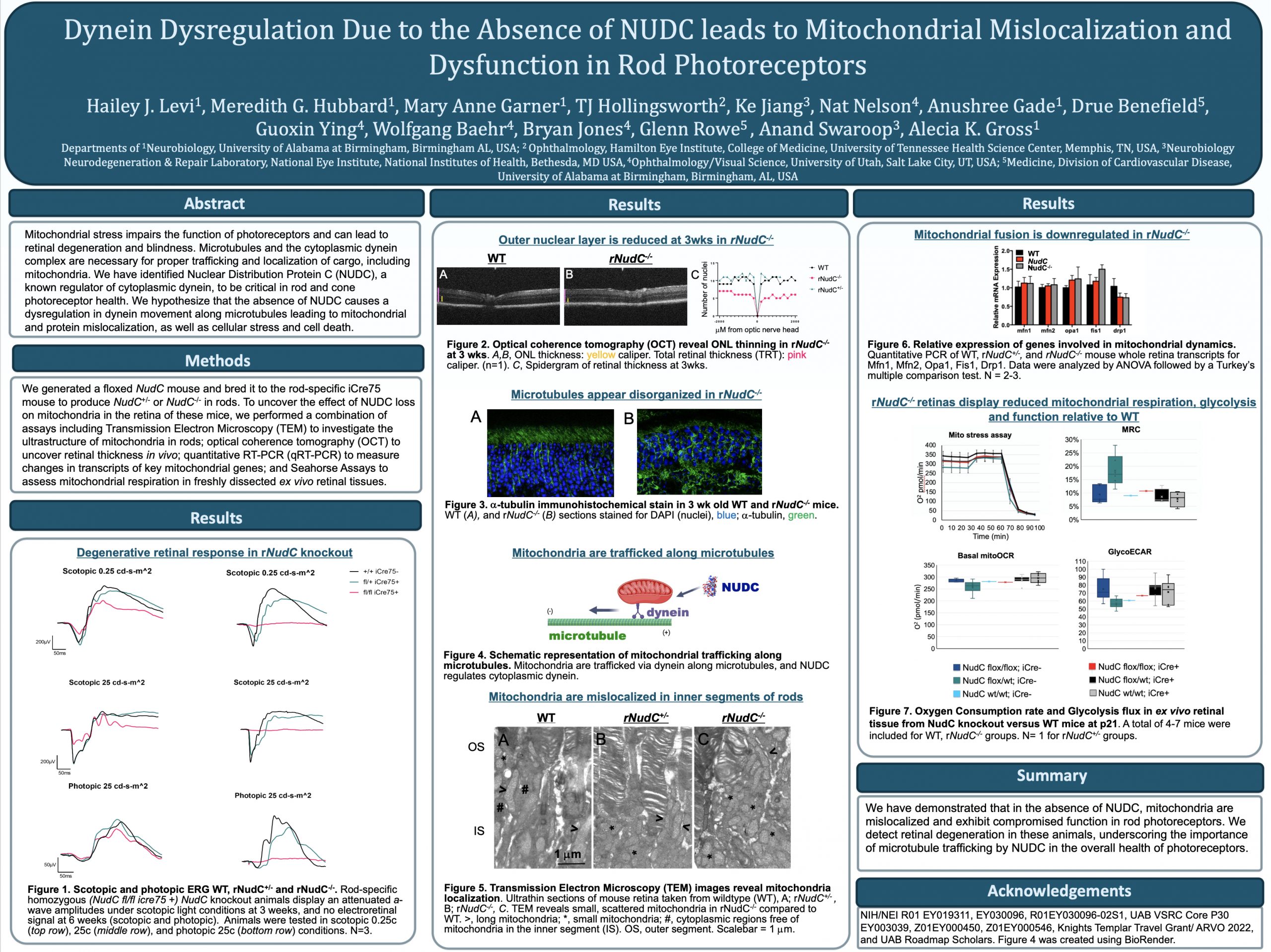
This abstract was presented today, May 4th at the 2022 Association for Research in Vision and Opthalmology (ARVO) meetings in Denver, Colorado by Hailey Levi @drpepperis100, Meredith Hubbard, Mary Anne Garner, TJ Hollingsworth, Ke Jiang, Nat Nelson, Anushree Gade, Drue Benefield, Guoxin Ying, Wolfgang Baehr, Bryan Jones@BWJones, Anand Swaroop, Glenn Rowe, and Alecia Gross @alecia144g.
Tag Archives: ARVO 2022
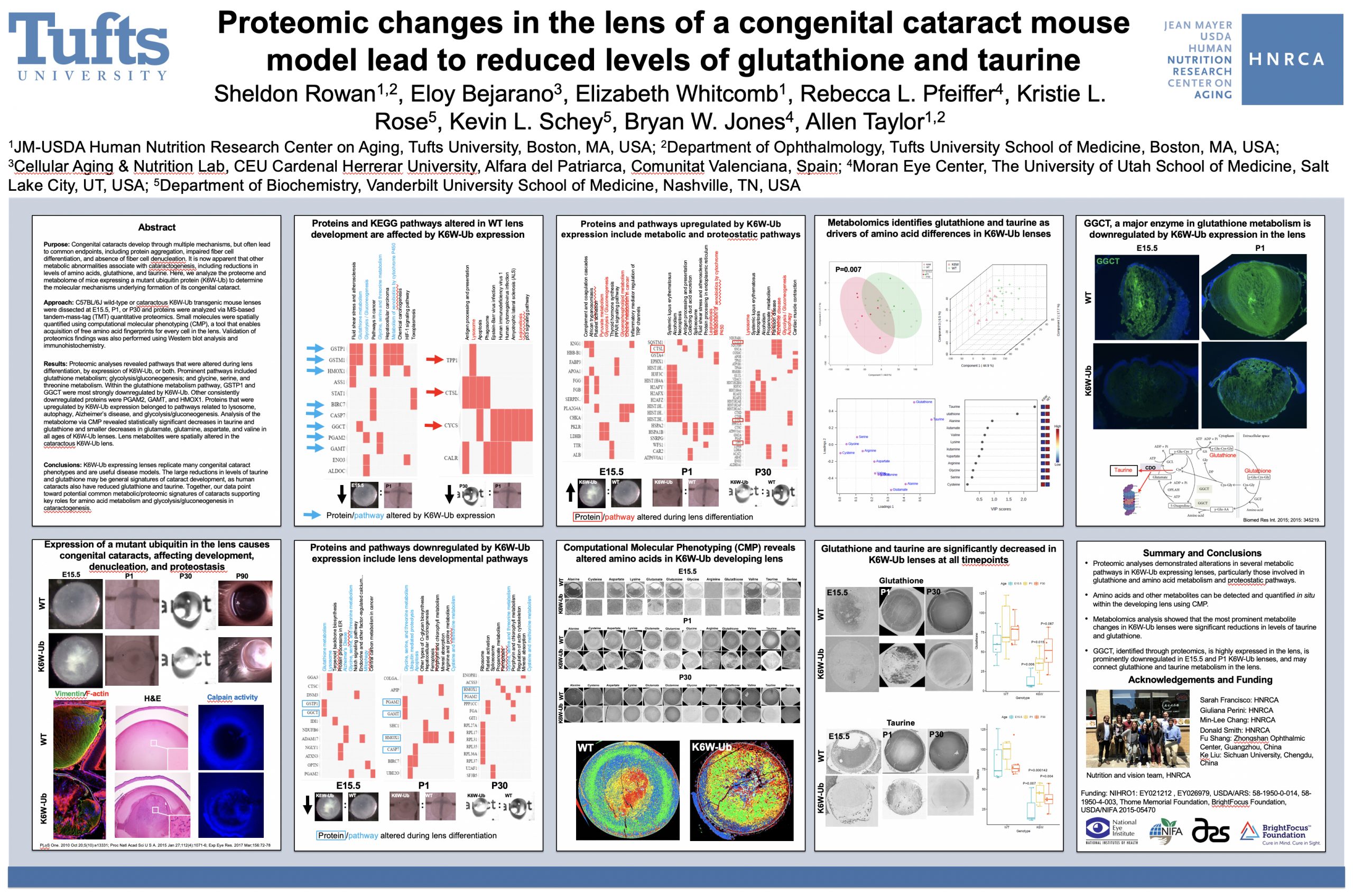
Proteomic changes in the lens of a congenital cataract mouse model lead to reduced levels of glutathione and taurine
This abstract was presented today, May 4th at the 2022 Association for Research in Vision and Opthalmology (ARVO) meetings in Denver, Colorado by Sheldon Rowan @SheldonRowan, Eloy Bejarano, Elizabeth Whitcomb, Rebecca Pfeiffer @BeccaPfeiffer19, Kristie Rose, Kevin Schey, Bryan Jones @BWJones, Allen Taylor.
Purpose: Congenital cataracts develop through multiple mechanisms, but often lead to common endpoints, including protein aggregation, impaired fiber cell differentiation, and absence of fiber cell denucleation. It is now apparent that other metabolic abnormalities associate with cataractogenesis, including reductions in levels of amino acids, glutathione, and taurine. Here, we analyze the proteome and metabolome of mice expressing a mutant ubiquitin protein (K6W-Ub) to determine the molecular mechanisms underlying formation of its congenital cataract.
Methods: C57BL/6J wild-type or cataractous K6W-Ub transgenic mouse lenses were dissected at E15.5, P1, or P30 and proteins were analyzed via MS-based tandem-mass-tag (TMT) quantitative proteomics. Small molecules were spatially quantified using computational molecular phenotyping (CMP), a tool that enables acquisition of free amino acid fingerprints for every cell in the lens. Validation of proteomics findings was also performed using Western blot analysis and immunohistochemistry.
Results: Proteomic analyses revealed pathways that were altered during lens differentiation, by expression of K6W-Ub, or both. Prominent pathways included glutathione metabolism; glycolysis/gluconeogenesis; and glycine, serine, and threonine metabolism. Within the glutathione metabolism pathway, GSTP1 and GGCT were most strongly downregulated by K6W-Ub. Other consistently downregulated proteins were PGAM2, GAMT, and HMOX1. Proteins that were upregulated by K6W-Ub expression belonged to pathways related to lysosome, autophagy, Alzheimer’s disease, and glycolysis/gluconeogenesis. Analysis of the metabolome via CMP revealed statistically significant decreases in taurine and glutathione and smaller decreases in glutamate, glutamine, aspartate, and valine in all ages of K6W-Ub lenses. Lens metabolites were spatially altered in the cataractous K6W-Ub lens.
Conclusions: K6W-Ub expressing lenses replicate many congenital cataract phenotypes and are useful disease models. The large reductions in levels of taurine and glutathione may be general signatures of cataract development, as human cataracts also have reduced glutathione and taurine. Key roles for amino acid metabolism and glycolysis/gluconeogenesis in cataractogenesis are emerging. Together our data point toward potential common metabolic/proteomic signatures of cataracts.
ARVO Mini-Symposium: Pathoconnectomics in Retinal Degeneration
Lab PI, Bryan Jones delivered a talk at the ARVO 2022 mini-symposium on Pathoconnectomics in Retinal Degeneration.
Abstract: Connectomics has demonstrated that synaptic networks and their topologies are precise and directly correlate with physiology and behavior. The next extension of connectomics is pathoconnectomics: to map neural network synaptology and circuit topologies corrupted by neurological disease in order to identify robust targets for therapeutics. The retina is ideal for pathoconnectomics approaches, and reveals common rules of how neural systems are wired, and how they break in neurodegenerative disease.