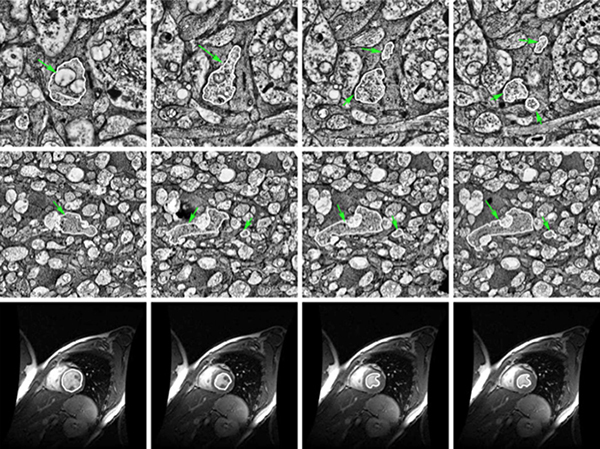
We have published another manuscript, Robust Segmentation based Tracking using an Adaptive Wrapper for Inducing Priors. This manuscript describes the work on adaptive tracing and proposes an algorithm that adapts a generic tracing algorithm to an application of interest. In our specific case, it is boundaries of cells in high frequency space in transmission electron microscopy images. But the approach in this paper is applicable to biological, medical, remote sensing and surveillance data as well utilizing priors specific to the application. The co-authors on the paper are: Vignesh Jagadeesh, James Anderson, Bryan W. Jones, Robert E. Marc, Steven K Fisher and B.S Manjunath.
Monthly Archives: September 2013
Retinal Connectomics: Toward Complete, Accurate Networks
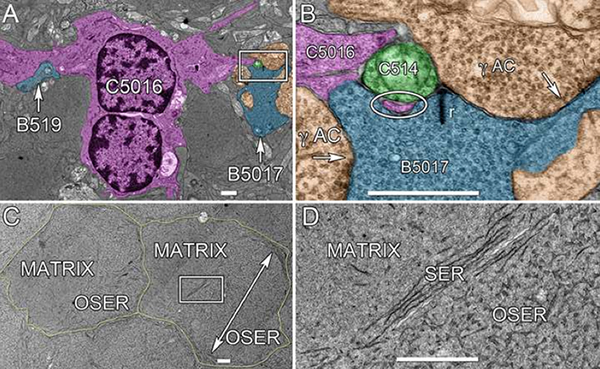
We have a new publication, Retinal connectomics: Toward complete, accurate networks in Progress in Retinal and Eye Research. Authors are: Robert E. Marc, Bryan W. Jones, Carl B. Watt, Crystal Sigulinsky, James R. Anderson and J. Scott Lauritzen.
Abstract:
Connectomics is a strategy for mapping complex neural networks based on high-speed automated electron optical imaging, computational assembly of neural data volumes, web-based navigational tools to explore 1012-1015 byte (terabyte to petabyte) image volumes, and annotation and markup tools to convert images into rich networks with cellular metadata. These collections of network data and associated metadata, analyzed using tools from graph theory and classification theory, can be merged with classical systems theory, giving a more completely parameterized view of how biologic information processing systems are implemented in retina and brain. Networks have two separable features: topology and connection attributes. The first findings from connectomics strongly validate the idea that the topologies complete retinal networks are far more complex than the simple schematics that emerged from classical anatomy. In particular, connectomics has permitted an aggressive refactoring of the retinal inner plexiform layer, demonstrating that network function cannot be simply inferred from stratification; exposing the complex geometric rules for inserting different cells into a shared network; revealing unexpected bidirectional signaling pathways between mammalian rod and cone systems; documenting selective feedforward systems, novel candidate signaling architectures, new coupling motifs, and the highly complex architecture of the mammalian AII amacrine cell. This is but the beginning, as the underlying principles of connectomics are readily transferrable to non-neural cell complexes and provide new contexts for assessing intercellular communication.
Seminar: Eduardo Rosa-Molinar, Towards Disentangling Adult Spinal Neural Circuits
Dr. Eduardo Rosa-Molinar will present a seminar at the Moran Eye Center at 4pm on October 8th, 2013 entitled Towards Disentangling Adult Spinal Neural Circuits. His talk will discuss the neurobiology of ultrafast spinal motor networks and imaging networks with field ion beam milling connectomics.
FASEB Bio-Art Competition Winner 2013
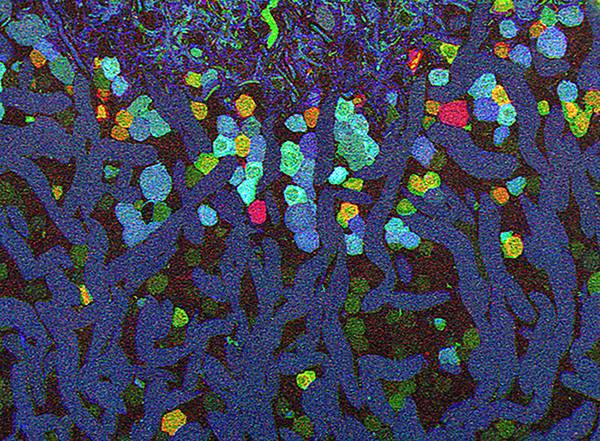
Bryan W. Jones and Robert E. Marc and were selected as 2013 FASEB BioArt Winners (Press release here). This image shows a region of an amazingly complex retina from a goldfish (Carassius auratus auratus) analyzed using tools called Computational Molecular Phenotyping (CMP) that reveal the metabolic state of the all cell types in tissues. These cells were labeled with antibodies for the presence of two fundamental amino acid metabolites (anti-glycine in red, anti-GABA in blue) and an amino acid tracer of physiologic activity (anti-AGB in green). These labels allow us to visualize the metabolic state and therefore, classes of bipolar, amacrine and horizontal cells. More details on the image here.