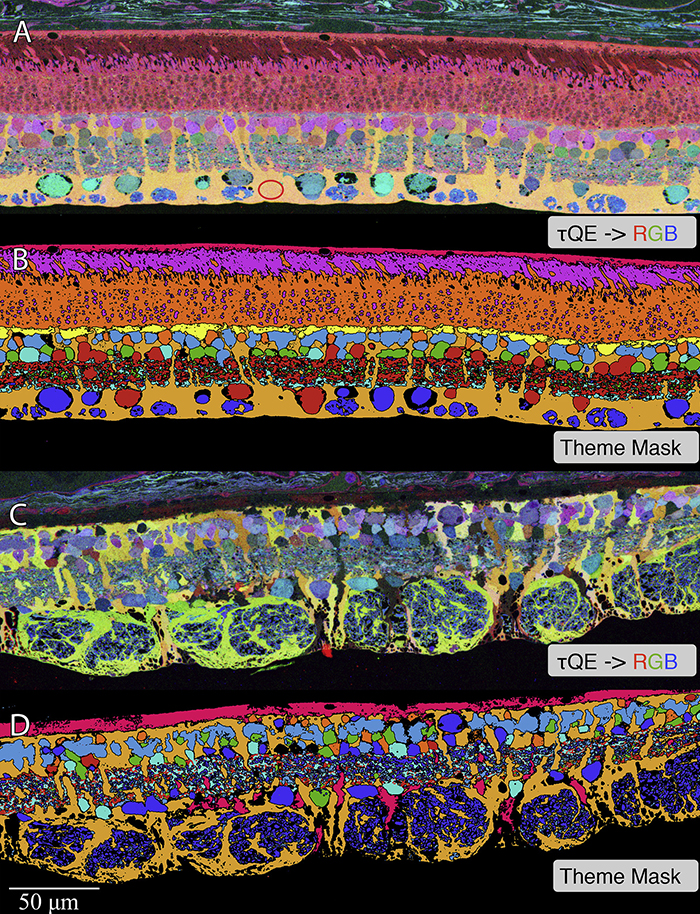
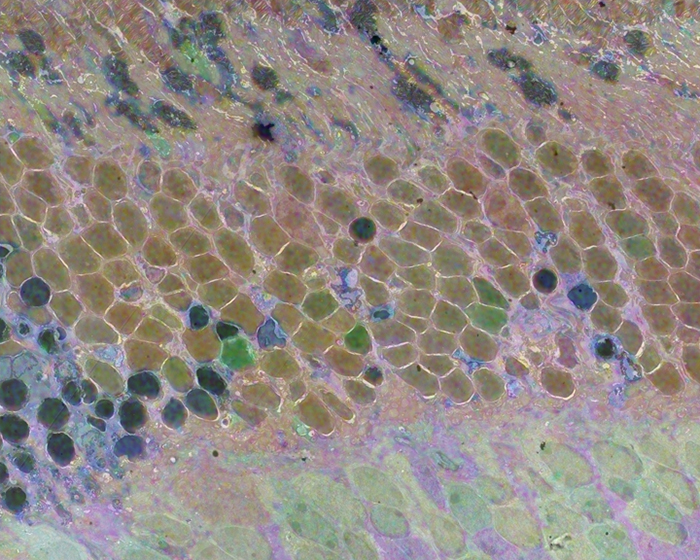
We have a new publication out (direct link, open access), Müller Cell Metabolic Chaos During Retinal Degeneration authored by Rebecca Pfeiffer, Robert Marc, Mineo Kondo, Hiroko Terasaki and Bryan W. Jones.
Abstract:
Müller cells play a critical role in retinal metabolism and are among the first cells to demonstrate metabolic changes in retinal stress or disease. The timing, extent, regulation, and impacts of these changes are not yet known. We evaluated metabolic phenotypes of Müller cells in the degenerating retina.
Retinas harvested from wild-type (WT) and rhodopsin Tg P347L rabbits were fixed in mixed aldehydes and resin embedded for computational molecular phenotyping (CMP). CMP facilitates small molecule fingerprinting of every cell in the retina, allowing evaluation of metabolite levels in single cells.
CMP revealed signature variations in metabolite levels across Müller cells from TgP347L retina. In brief, neighboring Müller cells demonstrated variability in taurine, glutamate, glutamine, glutathione, glutamine synthetase (GS), and CRALBP. This variability showed no correlation across metabolites, implying the changes are functionally chaotic rather than simply heterogeneous. The inability of any clustering algorithm to classify Müller cell as a single class in the TgP347L retina is a formal proof of metabolic variability in the present in degenerating retina.
Although retinal degeneration is certainly the trigger, Müller cell metabolic alterations are not a coherent response to the microenvironment. And while GS is believed to be the primary enzyme responsible for the conversion of glutamate to glutamine in the retina, alternative pathways appear to be unmasked in degenerating retina. Somehow, long term remodeling involves loss of Müller cell coordination and identity, which has negative implications for therapeutic interventions that target neurons alone.