We have a new manuscript from the lab in Experimental Eye Research, (PubMed link here). Metabolic changes and retinal remodeling in Heterozygous CRX mutant cats (CRXRDY/+). This manuscript is in collaboration with the Simon Petersen-Jones lab out of Michigan State University. Authors are: Laurence M. Occelli, Bryan W. Jones @BWJones, Taylor J. Cervantes, and Simon M. Petersen-Jones. The PDF is here.
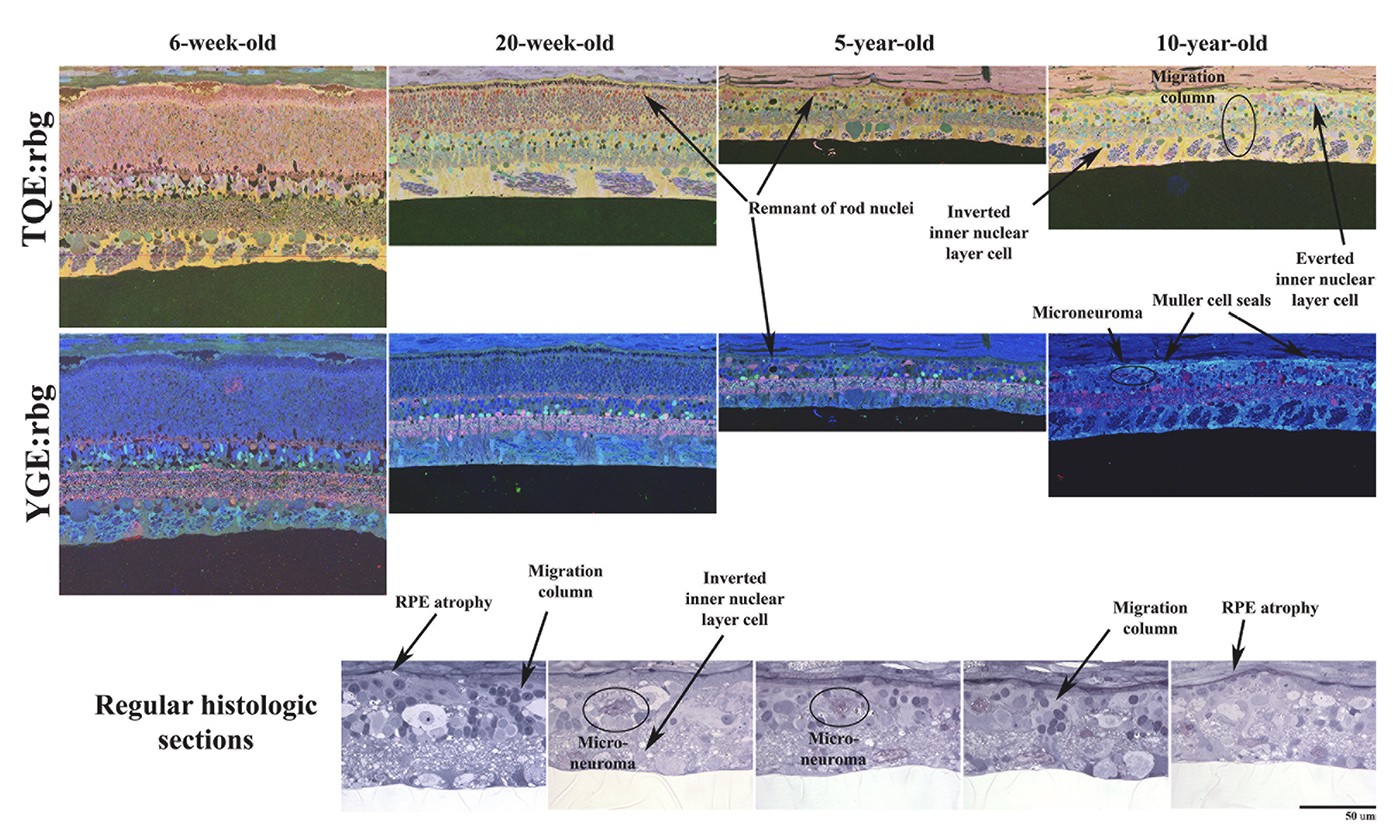
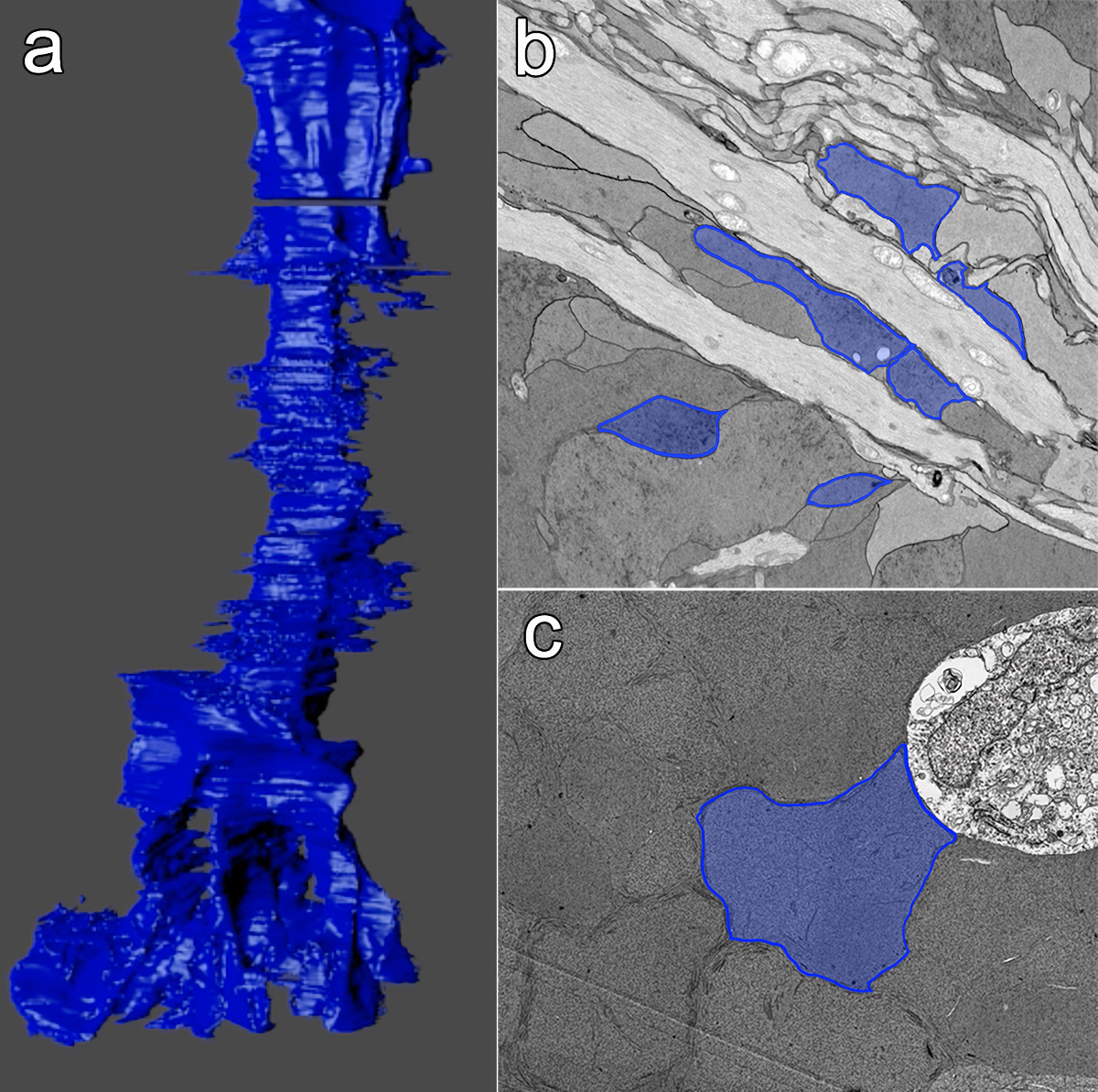
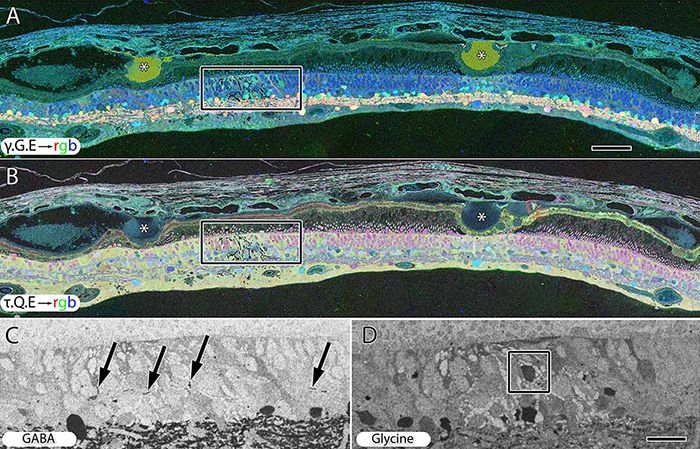
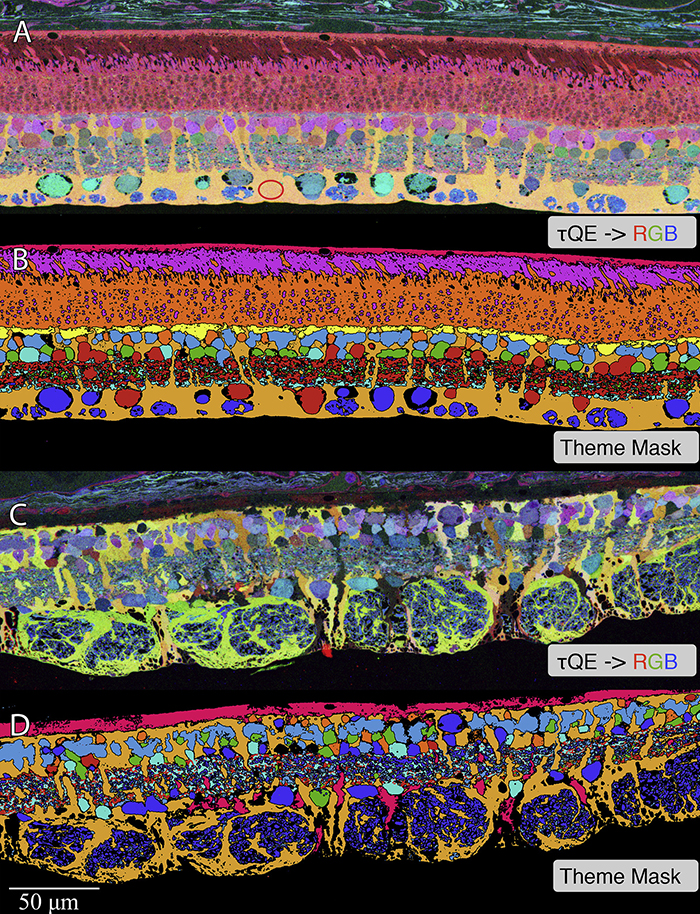
Abstract: CRX is a transcription factor essential for normal photoreceptor development and survival. The CRXRdy cat has a naturally occurring truncating mutation in CRX and is a large animal model for dominant Leber congenital amaurosis. This study investigated retinal remodeling that occurs as photoreceptors degenerate. CRXRdy/+ cats from 6 weeks to 10 years of age were investigated. In vivo structural changes of retinas were analyzed by fundus examination, confocal scanning laser ophthalmoscopy and spectral domain optical coherence tomography. Histologic analyses including immunohistochemistry for computational molecular phenotyping with macromolecules and small molecules. Affected cats had a cone-led photoreceptor degeneration starting in the area centralis. Initially there was preservation of inner retinal cells such as bipolar, amacrine and horizontal cells but with time migration of the deafferented neurons occurred. Early in the process of degeneration glial activation occurs ultimately resulting in formation of a glial seal. With progression the macula-equivalent area centralis developed severe atrophy including loss of retinal pigmentary epithelium. Microneuroma formation occurs in advanced stages as more marked retinal remodeling occurred. This study indicates that retinal degeneration in the CrxRdy/+ cat retina follows the progressive, phased revision of retina that have been previously described for retinal remodeling. These findings suggest that therapy dependent on targeting inner retinal cells may be useful in young adults with preserved inner retinas prior to advanced stages of retinal remodeling and neuronal cell loss.