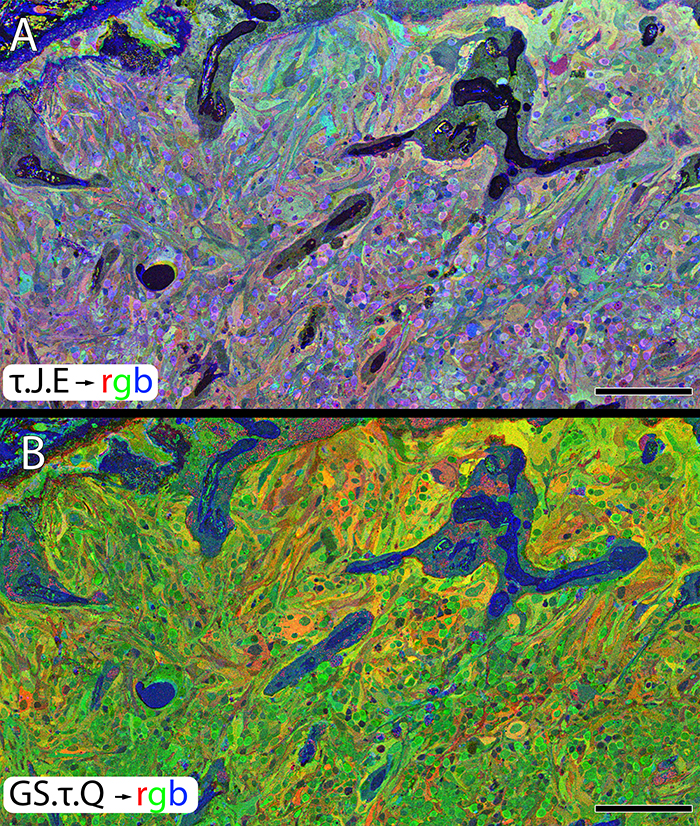
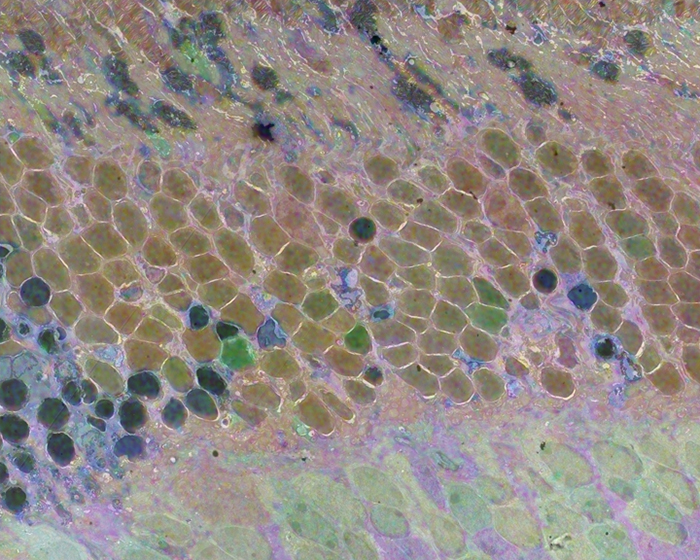
We have a new publication out (Direct Link, Free Open Access), Retinal Remodeling in Human Retinitis Pigmentosa authored by Bryan W. Jones, Rebecca Pfeiffer, Drew Ferrell, Carl Watt, Michael Marmor and Robert Marc.
Abstract: Retinitis Pigmentosa (RP) in the human is a progressive, currently irreversible neural degenerative disease usually caused by gene defects that disrupt the function or architecture of the photoreceptors. While RP can initially be a disease of photoreceptors, there is increasing evidence that the inner retina becomes progressively disorganized as the outer retina degenerates. These alterations have been extensively described in animal models, but remodeling in humans has not been as well characterized. This study, using computational molecular phenotyping (CMP) seeks to advance our understanding of the retinal remodeling process in humans. We describe cone mediated preservation of overall topology, retinal reprogramming in the earliest stages of the disease in retinal bipolar cells, and alterations in both small molecule and protein signatures of neurons and glia. Furthermore, while Müller glia appear to be some of the last cells left in the degenerate retina, they are also one of the first cell classes in the neural retina to respond to stress which may reveal mechanisms related to remodeling and cell death in other retinal cell classes. Also fundamentally important is the finding that retinal network topologies are altered. Our results suggest interventions that presume substantial preservation of the neural retina will likely fail in late stages of the disease. Even early intervention offers no guarantee that the interventions will be immune to progressive remodeling. Fundamental work in the biology and mechanisms of disease progression are needed to support vision rescue strategies.