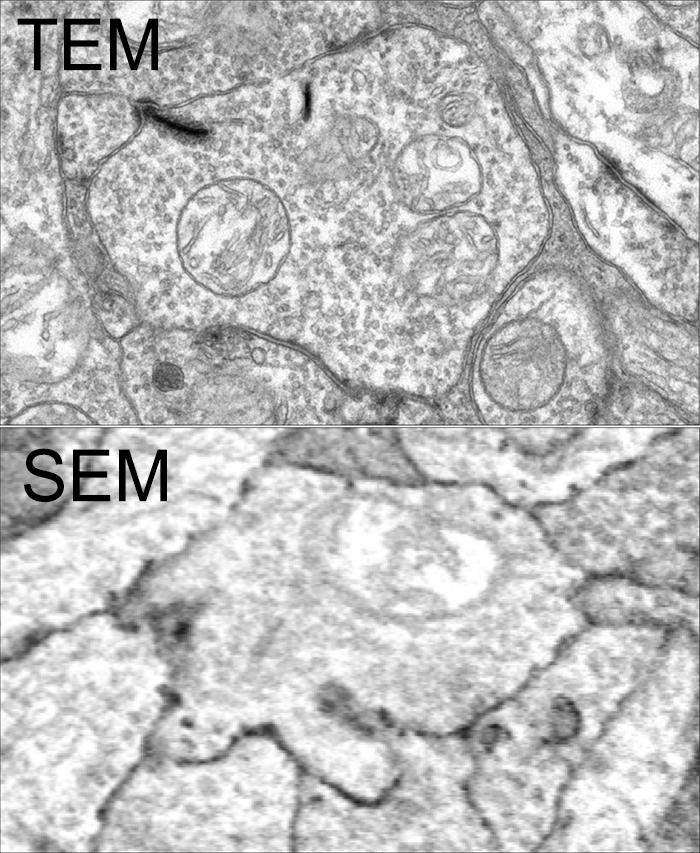
Super-resolution microscopy is a pretty big thing right now. But there is more than one way to get super-resolution microscopy results. There are a variety of approaches, most involving expensive new microscopes that preclude many scientists from participating in science that allows them to ask certain questions. However, if they have access to a standard transmission election microscope and have antibodies that are glutaraldehyde tolerant, they can participate and ask questions that allow them to get around some of the inherent limitations imposed by physics.
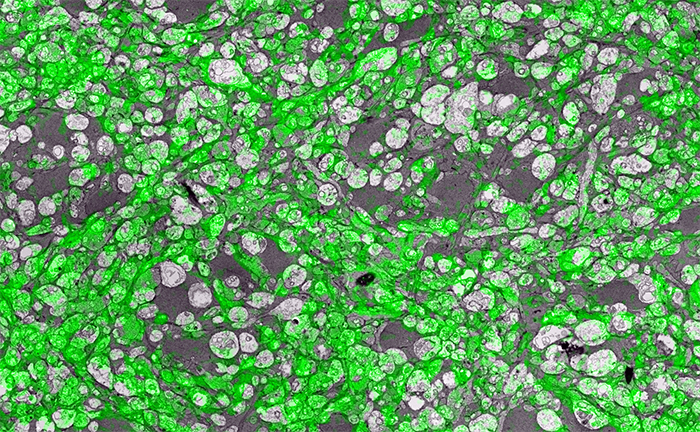
In the image above for example, we have GABA labeling in green superimposed upon ultrastructural data showing us *which* processes in the inner plexiform layer of the retina are GABAergic. Many of these processes are smaller than the wavelength of light.
There are multiple ways to get here of course with some very expensive microscopes offering dual light and electron microscopy approaches and yet other microscopes offering purely optical based solutions. However, this is cheap and easy and accessible to many with the basic electron microscopy resources. Robert Marc first used this approach in back in 2000, and we subsequently used it for quite a bit of work for my Ph.D. dissertation in 2003, and notably in this paper. It is also an integral technique associated with our connectomics efforts.
That said, I’ll need at some point soon to find the resources to get a traditional optical super-resolution microscopy solution to answer some questions we have in the lab on neural degenerative disease.